Demonstrations¶
Busbar¶
“Electrical Heating in a Busbar” is an example model used in the tutorial in “Introduction to Comsol Multiphysics” and explained there in great detail. The section “Getting the Maximum and Minimum Temperature” demonstrates how two obtain the two temperature extremes within the Comsol GUI.
The following Python code does the same thing programmatically:
import mph
client = mph.start()
model = client.load('busbar.mph')
model.solve()
(x, y, z, T) = model.evaluate(['x', 'y', 'z', 'T'])
(Tmax, Tmin) = (T.max(), T.min())
(imax, imin) = (T.argmax(), T.argmin())
print(f'Tmax = {T.max():.2f} K at ({x[imax]:5f}, {y[imax]:5f}, {z[imax]:5f})')
print(f'Tmin = {T.min():.2f} K at ({x[imin]:5f}, {y[imin]:5f}, {z[imin]:5f})')
This outputs the exact same numbers that appear in the table of the GUI:
Tmax = 330.42 K at (0.105000, -0.024899, 0.053425)
Tmin = 322.41 K at (0.063272, 0.000000, 0.000000)
You could now sweep the model’s parameters, for example the length L
or width wbb of the busbar.
Compacting models¶
We usually save models to disk after we have solved them, which includes the solution and mesh data in the file. This is convenient so that we can come back to the model later, but don’t have to run the simulation again, which may take a long time. However, the files then require a lot of disk space. After a while, we may want to archive the models, but trim the fat before we do that.
To compact all model files in the current working directory, we can do this:
import mph
from pathlib import Path
client = mph.start()
for file in Path.cwd().glob('*.mph'):
print(f'{file}:')
model = client.load(file)
model.clear()
model.save()
The script compact_models.py in the “demos” folder of the
source-code repository is a refined version of the above code. It
displays more status information and also resets the modeling history.
Note that we could easily go through all sub-directories recursively
by replacing glob with rglob. However, this should be used with
caution so as to not accidentally modify models in folders that were
not meant to be included.
Multiple processes¶
As explained in Limitations, we cannot run more than one Comsol session inside the same Python process. But we can start multiple Python processes in parallel, thanks to the multiprocessing module that is part of the standard library.
So, other than MPh itself, we are going to need multiprocessing,
as well as queue, also from the standard library, though only for
the queue.Empty exception type that it provides.
import mph
import multiprocessing
import queue
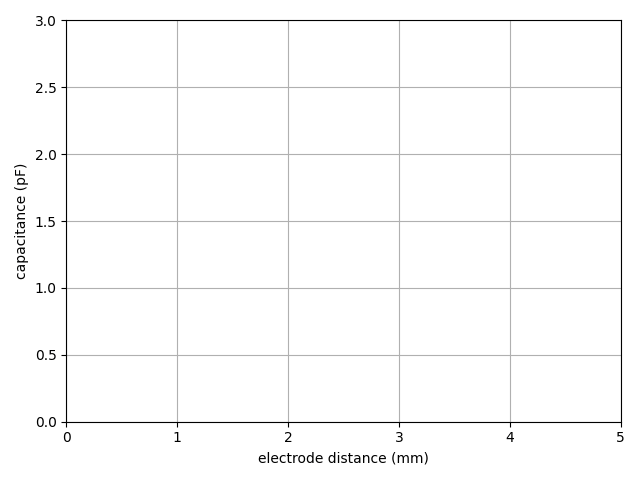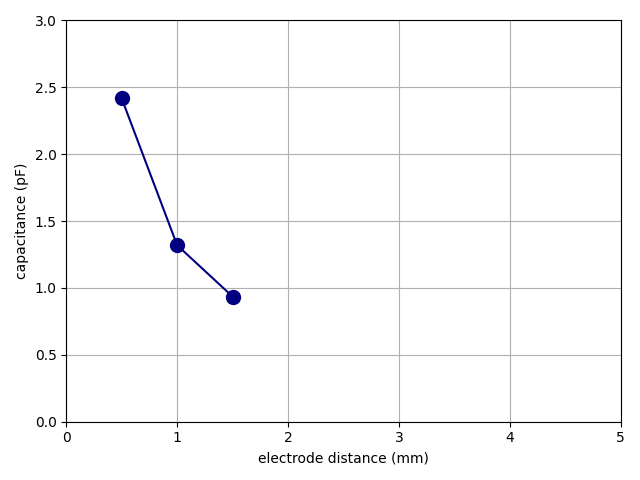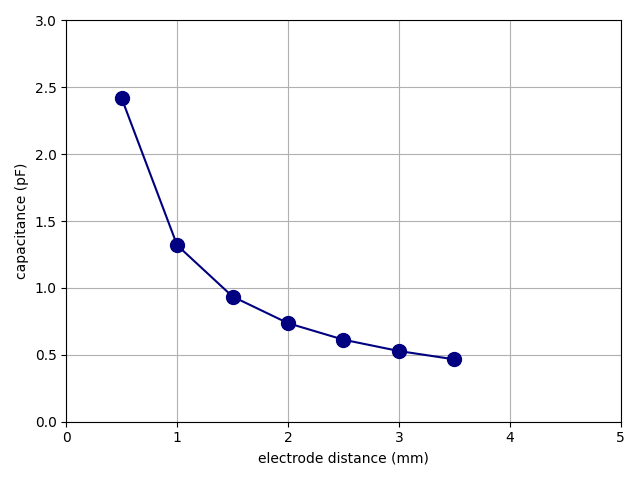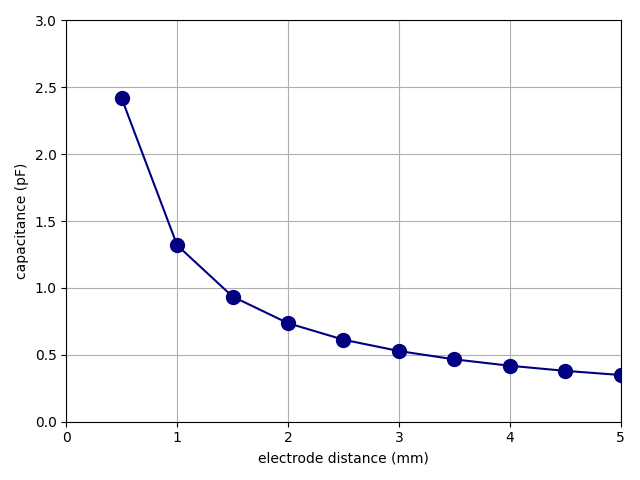
In this demonstration, we will solve the model capacitor.mph from the
“tests” folder of the source-code repository, the same model
we used in the Tutorial. We want to sweep the electrode
distance d and calculate the capacitance C for each value of the
distance, ranging from 0.5 to 5 mm.
values = [0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0]
Next, we define the function that we intend to run in every process,
i.e. the “worker”. The function sets up the Comsol session when the
process starts, then keeps solving the model for every distance value
that it receives via a jobs queue. Each time, it evaluates the
solution and returns the capacitance via a results queue. It does
so until the jobs queue is exhausted, upon which the function
terminates, and with it Comsol session and Python process.
def worker(jobs, results):
client = mph.start(cores=1)
model = client.load('capacitor.mph')
while True:
try:
d = jobs.get(block=False)
except queue.Empty:
break
model.parameter('d', f'{d} [mm]')
model.solve('static')
C = model.evaluate('2*es.intWe/U^2', 'pF')
results.put((d, C))
Each worker will only use one of the processor cores available on the machine, as that’s the whole point: We want to achieve maximum speed-up of, say, a parameter sweep, by having each core work on a job corresponding to one of the many parameter values, which it can do independently of work being done for any other value.
We could also solve this sequentially, one parameter value at a time. Comsol’s solver could then make use of all cores and would also employ some kind of parallelization in its internal computation. But the speed-up would not scale linearly with the number of cores, especially for large numbers of them.
We might use a “parametric sweep”, a feature that Comsol does offer. But by doing this in Python we retain full programmatic control of which parameter is solved for and when. The parameter values don’t have to be hard-coded, they could come from user input or be generated depending on the outcome of previous simulations. For example, this approach lends itself to iterative optimization schemes such as the “genetic algorithm”, where a batch of simulations would be run for each new “generation”.
Note how the returned results also contain the input parameter. As the worker processes will run asynchronously in parallel, we cannot take for granted that output is returned in input order.
Before we start the computation, we add all parameter values to the
jobs queue:
jobs = multiprocessing.Queue()
values = [0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0]
for d in values:
jobs.put(d)
We also have to provide the results queue, which is of course empty
at first.
results = multiprocessing.Queue()
Then we can start a number of workers, say four:
for _ in range(4):
process = multiprocessing.Process(target=worker, args=(jobs, results)
process.start()
It may be a good idea to hold on to the process objects and add them
to a list processes, just so that Python’s garbage collection won’t
accidentally delete them while the external processes are running.
Finally, still in the main process that starts all the workers, we can
collect the results. We use a for loop and exploit the fact that
there will be as many results as there were jobs to begin with.
for _ in values:
(d, C) = results.get()
We would then display them, plot them, save them to a file, or whatever it is we do with simulation results.
The complete script worker_pool.py, which implements all of the above
and also irons out some wrinkles not covered here for the sake of
brevity, can be found in the “demos” folder of the source-code
repository. As it runs, it displays a live plot such as the one that
follows. It is reproduced here preserving the real time from a run with
two workers. Observe how the first two data points do in fact come in
out of order.
A more advanced implementation may use a class derived from multiprocessing.Process instead of a mere function, just to be able
so save state. For long-running simulations it would make sense to
store jobs and results on disk, rather than in memory, so that the
execution of the queue may be resumed after a possible interruption.
In that case, one may, or may not, find the subprocess
module more convenient for starting the external processes. The
worker implementation would then be in a separate module that is run
as a script.
Creating models¶
While it is not the primary focus of MPh to create model features and change settings, it is however possible — thanks to JPype, the Python-to-Java bridge that underpins MPh. All credit to the JPype developers for making this possible.
Let’s take this Comsol blog entry as an example: Automate Your Modeling Tasks with the Comsol API for use with Java. It starts with the following Java code example:
import com.comsol.model.*;
import com.comsol.model.util.*;
public class HelloWorld {
public static void main(String[] args) {
run();
}
public static Model run() {
Model model = ModelUtil.create("Model");
model.modelNode().create("comp1");
model.geom().create("geom1", 3);
model.geom("geom1").feature().create("blk1", "Block");
model.geom("geom1").feature("blk1").set("size", new String[]{"0.1", "0.2", "0.5"});
model.geom("geom1").run("fin");
return model;
}
}
What it does is, it creates a model, which contains a 3d geometry component that is just a block 0.1 by 0.2 by 0.5 meters in size.
In Python, we would achieve the same with the code that follows. It
uses the .java attribute of the Python wrapper to access the model’s
Java API directly.
import mph
client = mph.start()
pymodel = client.create('model')
model = pymodel.java
model.modelNode().create("comp1");
model.geom().create("geom1", 3);
model.geom("geom1").feature().create("blk1", "Block");
model.geom("geom1").feature("blk1").set("size", ["0.1", "0.2", "0.5"]);
model.geom("geom1").run("fin");
Note how the functional Java code (excluding syntax sugar) was
essentially copied and pasted, even the semicolons, which Python
simply ignores. We had to replace new String[]{"0.1", "0.2", "0.5"}
because Python does not know what new means. There, Java expects a
list of three strings. So we replaced the expression with
["0.1", "0.2", "0.5"], the Python equivalent of just that: a list
of these three strings.
Occasionally, when translating Java (or Matlab) code you find in the documentation — or a blog entry, as the case was here —, you will have to amend code lines such as the one above. But they are few and far between. And the error messages you might receive would point you in the right direction.
The upside of using the Python interface is:
You don’t really need to know Java. Just a little, to understand that occasionally we have to take care of type conversions that JPype cannot handle all by itself. Which is rare.
You don’t need to install Java. It just ships with Comsol. You also don’t need to bother with compiling Java source code to Java classes via
comsolcompile.You can use Python introspection to understand how Comsol models are “created in code”. This may be the most productive feature. The Comsol documentation explains a lot of things, but not every little detail. The function
mph.inspect()makes introspection even easier, as it formats the output more nicely than Python’s built-indir().
Finally, to save the model created in the above example, we can just do this:
pymodel.save('model')
This stores a file named model.mph in the working directory, which
we can then open in the Comsol GUI or use in any other Python, Java,
or Matlab project.